Examples¶
This page gives a brief example for a full run of then main components of SyConn.
The corresponding code is available in /examples/full_run_example.py and
the example data with all pre-trained models can be found in SyConnDenseCube(.zip) (go to our Website for Download).
Running the example¶
Given that the example zipfile was extracted, running the analysis would just be:
python2 SyConn/examples/full_run_example.py </path/to/example_folder> [--gpus {None | <int> <int> ...}, --CNNsize {1 | <int>}, --qsub_pe {None | <str>}, --qsub_queue {None | <str>}]
By default the CNN prediction uses only CPUs (None option; up to as many cores as specified in the ~/.theanorc). GPUs can be made available by adding their ids (specified in nvidia-smi). The CNNsize argument allows the adjustment of the memory used on each GPU, where 4 is the highest possible value (more VRAM used) and 0 the smallest value. Defining either a qsub queue or qsub parallel environment lets the non-CNN part of the pipeline making use of a qsub cluster.
Data Set¶
This data set is a subset of the zebra finch area X dataset j0126 acquired by Jörgen Kornfeld.
The above figure shows cell tracings in 3D EM data cube, which is the required data to perform automated structural analysis using SyConn. The following section is a walk-through of all major steps.
Applying SyConn – step by step¶
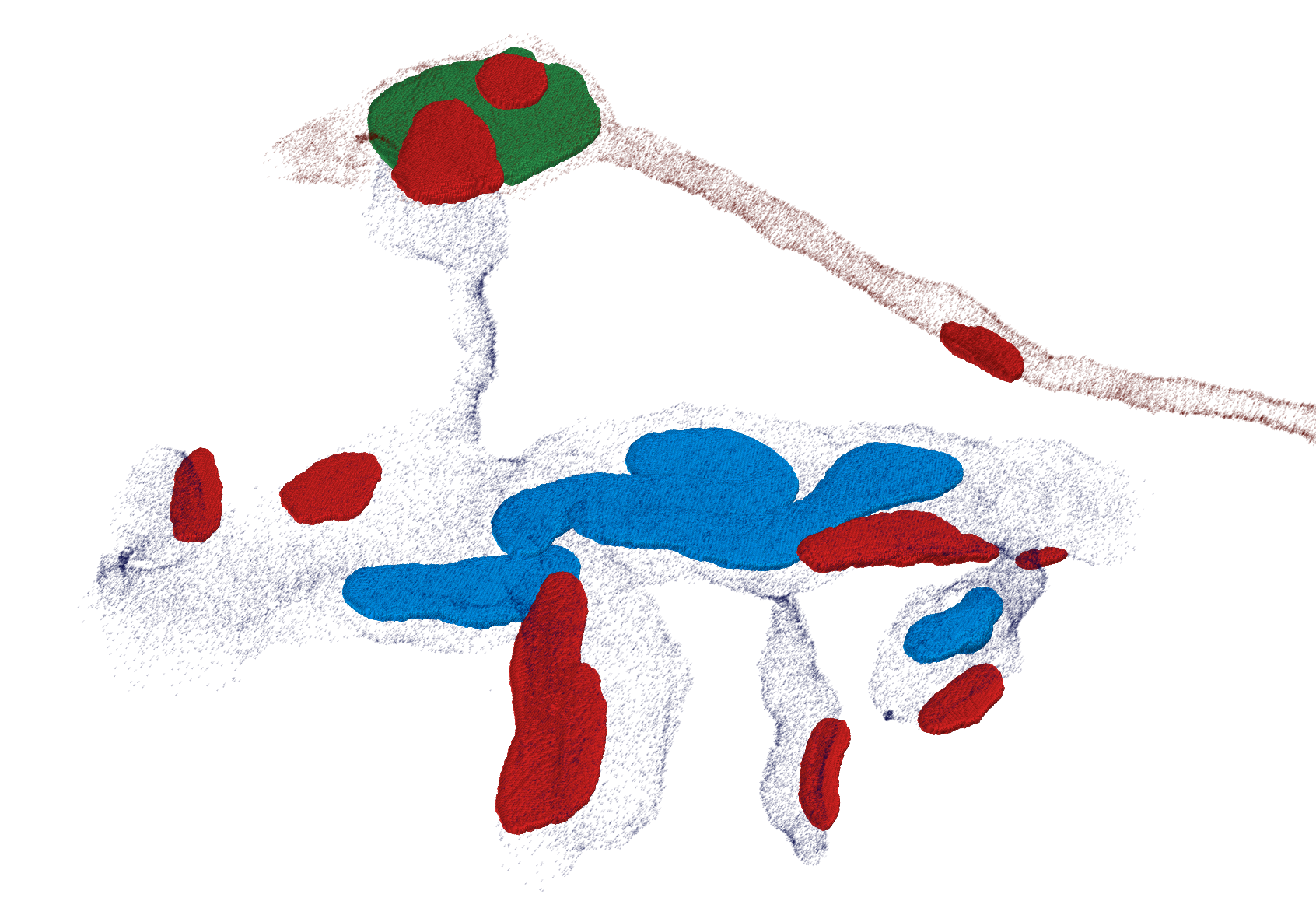
At first, membrane and intracellular space is predicted as barrier (black) and cell components, such as mitochondria (blue), vesicle clouds (green) and synaptic junctions (red) are inferred using a CNN with a 3D perceptive field of view (image below). Additionally, another 3D CNN predicts symmetric and asymmetric synaptic junction regions which get combined with the predicted synaptic junctions later on.
Thresholding and connected components analysis then reveal volumetric objects for each class. Based on the cell tracings and the barrier prediction a cell hull sampling is performed which enables the mapping of these objects to individual cells (embedding tracings in biological environment). Furthermore, contact sites of adjacent cell pairs can be distinguished from synapses combining properties of corresponding contact site and, if existent, synaptic junction.
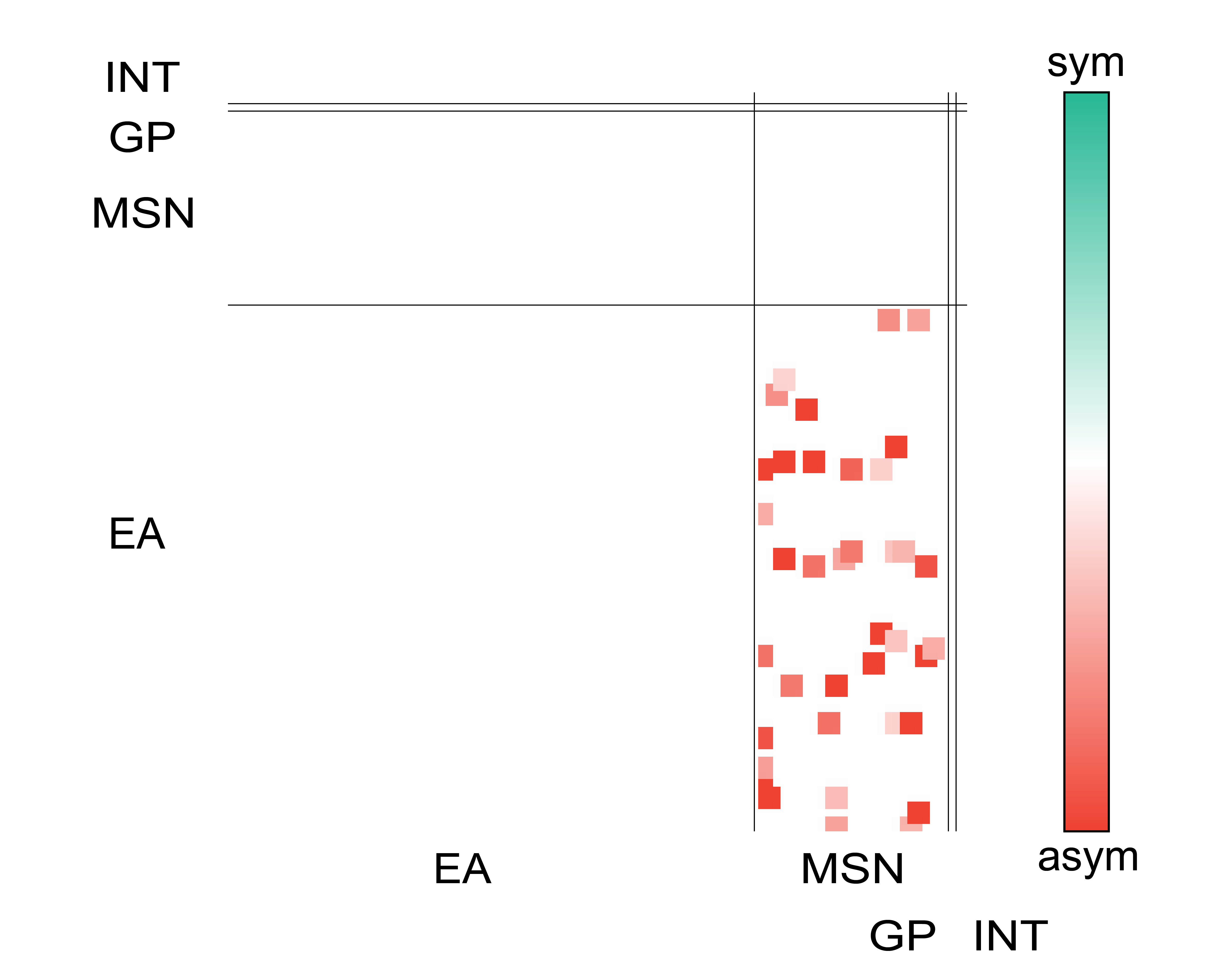
With these priors sub-cellular compartments can be inferred (axon/dendrite/soma and spines in dendrites) to further assign types (EA, MSN, GP, INT) to every cell and create a wiring diagram combining knowledge about pre and post synapses (sub-cellular compartments), area of synaptic junctions and cell functions.